Small GTPase OsRac1 in plant immunity
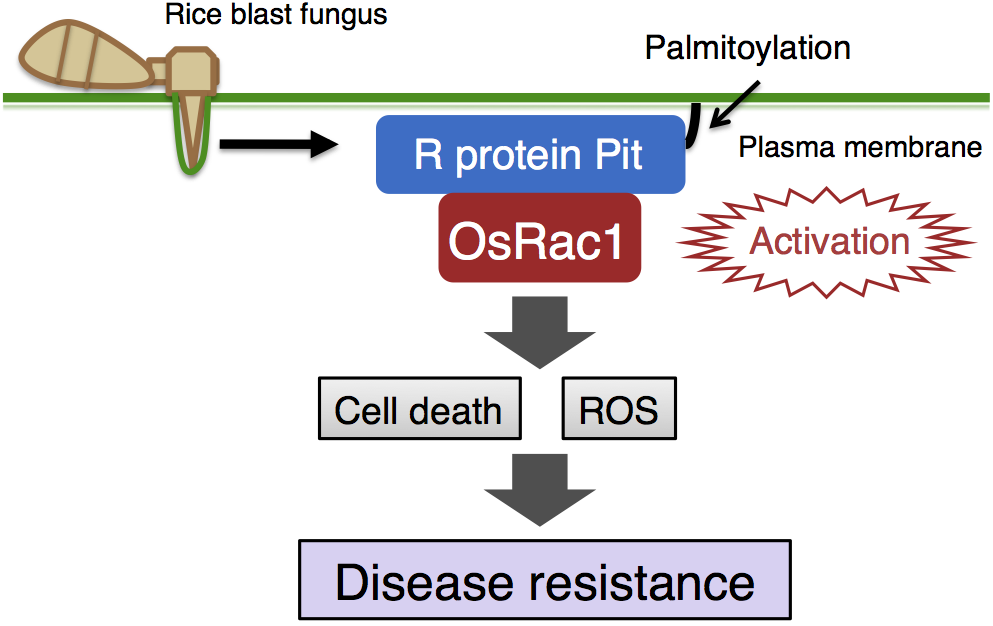
A master regulator of immunity, OsRac1, and R protein signaling
Resistance (R) proteins are crucial
intracellular receptors that detect attack by insects and invasion by
various pathogens including fungi, bacteria and viruses. However, the
signaling molecules which mediate R protein-induced immune responses
are not yet fully understood. We have previously shown that an
intracellular switch, the small GTPase OsRac1, is a master regulator
controlling immunity in rice (Kawano et al., 2010b, Kawano and
Shimamoto, 2013). However, the mechanism by which OsRac1 receives
signals from the immune receptors and becomes activated has remained
unclear. Thus, we explored OsRac1-binding proteins and identified the R
protein Pit, which is a immune receptor for rice blast
fungus, a prominent microbial disease of rice (Kawano et al., 2010a).
Through various analyses, we demonstrated that OsRac1 functions as a
molecular switch, controlling ROS production and hypersensitive cell
death downstream of Pit. In addition, we have found that
anchoring Pit to the plasma membrane through palmitoylation, a type
of lipid modification, is required for Pit-induced activation of
OsRac1 on the plasma membrane (Kawano et al., 2014). Our work has
therefore revealed the signaling pathway of the R protein Pit through
OsRac1.
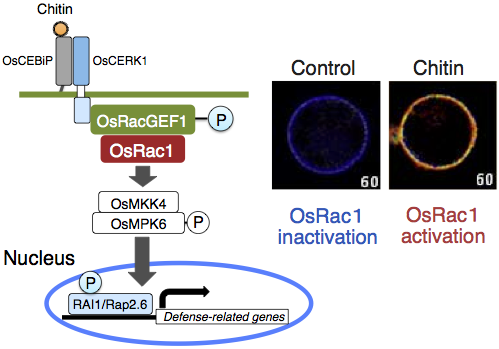
In vivo monitoring of OsRac1 activity and chitin-triggered immunity
Given that the small GTPase OsRac1 is a master regulator controlling rice immunity, monitoring its activation within plant cells was believed to be the next key step in understanding plant immunity. By creating a bio-imaging sensor using the Forster resonance energy transfer (FRET) of fluorescent proteins, we were the first to successfully monitor the activation of a plant small GTPase in vivo (Kawano et al., 2010a). Using this sensor, we identified the R protein Pit as an immune receptor that activated OsRac1 on the plasma membrane (Kawano et al., 2010a), and observed that OsRac1 was activated within 3 min after sensing chitin, a cell wall component of rice blast fungus (Akamatsu et al., 2013). Furthermore, we revealed that the signal from the chitin receptor OsCERK1 was transmitted to OsRac1 through the OsRac1 activator protein OsRacGEF1. We have also reported that OsRac1 controls the expression of defense-related genes through the MAPK OsMPK6 and the transcription factors RAI1 and Rap2.6 (Kim et al., 2012, Wamaitha et al., 2012).
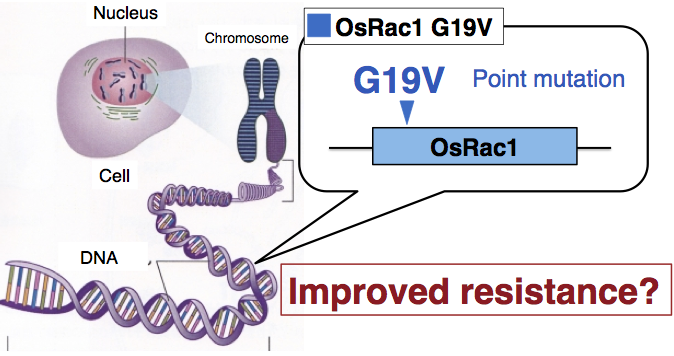
Homologous recombination-based gene targeting of OsRac1 to enhance rice immunity
Overexpression
of defense-related
proteins often causes stress and results in a reduction of biomass.
Thus, technological innovation has been considered essential to provide
plants with improved resistance to diseases while maintaining biomass.
Rice transformed with constitutively active OsRac1 G19V, which
was overexpressed using the 35S promoter that allows constant high
expression, displays improved resistance against pathogens, but a
reduced yield of rice because of a reduction in fertility. Therefore,
rather than utilizing conventional gene transfer methods, we applied a
gene-targeting method that uses homologous recombination, developed by
Dr. R. Terada of Meijo University. Using this method, we introduced
a point mutation that makes the endogenous OsRac1 gene constitutively
active and investigated whether disease resistance in rice could be
improved, thereby avoiding agriculturally unfavorable characters
(Dang et al., 2013). Because selection markers can be removed through
site-specific recombination, the gene-targeting method utilized in this study is a
groundbreaking, marker-free genetic engineering technique with the
ability to modify targeted genes as closely as possible to natural
mutations. Introduction of the constitutively active mutation into the
OsRac1 gene caused an increase
in the expression of transcription
factors such as WRKY, and of
defense-related genes including PAL1
and
PBZ1. A resistance response
similar to hypersensitive cell death was
also observed at the site of rice blast fungus infection.